Publications
Highlights
The Architecture of SARS-CoV-2 Transcriptome
SARS-CoV-2 is a betacoronavirus responsible for the COVID-19 pandemic. We have sequenced both genomic and subgenomic RNAs to understand the transcriptional landscape of the viral activities. Two complementary techniques were adopted for this study. DNA nanoball sequencing shows that the transcriptome is highly complex, owing to numerous discontinuous transcription events. SARS-CoV-2 produces transcripts encoding unknown ORFs with fusion, deletion, and frameshift in addition to the canonical genomic and nine subgenomic RNAs. Using nanopore direct RNA sequencing, we mapped the primary structures of full-length subgenomic RNAs with discontinuities. The relatively new technique also enables the analysis of RNA modifications. We found frequent modifications that are specific to a subset of viral RNAs. Functional investigation of the unknown transcripts and RNA modifications discovered in this study will open new directions to our understanding of the life cycle and pathogenicity of SARS-CoV-2.
D. Kim, J.-Y. Lee, J.-S. Yang, J. W. Kim, V. N. Kim, and H. Chang
Cell, 181(4):914-921.e10
Terminal Uridylyltransferases Execute Programmed Clearance of Maternal Transcriptome in Vertebrate Embryos
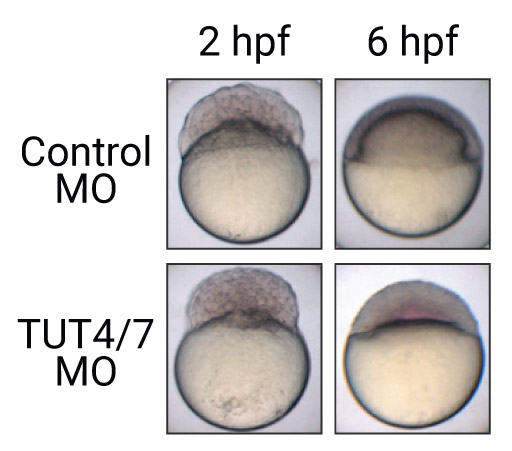
Life starts with the RNAs and proteins derived from its mother in animals. For the first few cell cycles, maternally deposited RNAs are precisely used to produce the right amount of the proteins at the right place. Once the bootstrapping of embryo finishes, the maternal RNAs are replaced by the zygotically transcribed RNAs. In this paper, we surveyed the poly(A) tails during the earliest cell cycles of vertebrate embryos to understand how the transition is accurately controlled. A terminal uridylyltransferase, TUT7, is found to be a driving factor initiating maternal RNA degradation. The preference to short poly(A) tails of TUT7 contributes to the selectivity of the process. This paper demonstrates the unique opportunities of TAIL-seq in the studies of biological phenomena coming from its unprecedented throughput and resolution in contrast to the existing biochemical methods.
H. Chang1, J. Yeo1, J.-g. Kim, H. Kim, J. Lim, M. Lee, H. H. Kim, J. Ohk, H.-Y. Jeon, H. Lee, H. Jung, K.-W. Kim, and V. N. Kim
Molecular Cell, 70(1):72–82
Uridylation by TUT4 and TUT7 Marks mRNA for Degradation
During the development of TAIL-seq, we found that the mammalian poly(A) tails often carry additional uridines at the 3′ end. What do those U tails do? We sought the enzymes responsible for the uridylation of poly(A) tails. TUT4 and TUT7 were found to be working for it redundantly. Without U tails, messenger RNAs had significantly longer half-lives. Which step in the mRNA catabolic process does the U tailing kicks in? Through the comparisons of tail profiles in dozens of perturbation conditions, we conclude that it happens after deadenylation and both 5′→3′ and 3′→5′ decay pathways are facilitated by uridylation of poly(A) tails. These discoveries were possible particularly thanks to the ability of TAIL-seq to focus in the oligo(A) tails apart from long poly(A) tails.
J. Lim1, M. Ha1, H. Chang1, S. C. Kwon, D. K. Simanshu, D. J. Patel, and V. N. Kim
Cell, 159(6):1365–1376
TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications
Eukaryotic messenger RNAs have long tails. They are long homogenous stretches comprised of only adenosine residues. It is widely known that these poly(A) tails protect RNAs from degradation and enhances the translation. Despite the 40 years of researches on the poly(A) tails, it was unclear how different are the poly(A) lengths in different genes and the detailed sequence composition of poly(A) tails. The major obstacle in adopting modern high-throughput techniques for poly(A) analysis was the long homopolymeric sequence. The second-generation sequencers such as Illumina SBS and IonTorrent can’t deal with such long monotonous repeats due to failure of cumulative error estimation (Illumina) or signals exceeding the dynamic range (IonTorrent). To overcome the status, we had a look into the raw images directly from the fluorescence microscope equipped in the Illumina sequencers. We could find a way to recover the poly(A) tail length from the distorted fluorescence signals. Finally, we developed a high-throughput technique profiling poly(A) tail length distributions as well as 3′-end modifications of thousands of mRNAs simultaneously. The software is available from our GitHub repository.
H. Chang1, J. Lim1, M. Ha, and V. N. Kim
Molecular Cell, 53(6):1044–1052
Full List
Viral Hijacking of the TENT4–ZCCHC14 Complex Protects Viral RNAs via Mixed Tailing
D. Kim, Y.-S. Lee, S.-J. Jung, J. Yeo, J. J. Seo, Y.-Y. Lee, J. Lim, H. Chang, J. Song, J. Yang, J.-S. Kim, G. Jung, K. Ahn, and V. N. Kim
Nature Structural & Molecular Biology
(2020), 27:581–588
‣
The Architecture of SARS-CoV-2 Transcriptome
D. Kim, J.-Y. Lee, J.-S. Yang, J. W. Kim, V. N. Kim, and H. Chang
Cell
(2020), 181(4):914–921.e10
Development of a Laboratory-safe and Low-cost Detection Protocol for SARS-CoV-2 of the Coronavirus Disease 2019 (COVID-19)
J. Won, S. Lee, M. Park, T. Y. Kim, M. G. Park, B. Y. Choi, D. Kim, H. Chang, V. N. Kim, and C. Justin Lee
Experimental Neurobiology
(2020), 29(2):107–119
Bias-Minimized Quantification of MicroRNA Reveals Widespread Alternative Processing and 3′ End Modification
H. Kim, J. Kim, K. Kim, H. Chang, K. You, and V. N. Kim
Nucleic Acids Research
(2019), 47(5):2630–2640
Mixed Tailing by TENT4A and TENT4B Shields mRNA from Rapid Deadenylation
J. Lim, D. Kim, Y. Lee, M. Ha, M. Lee, J. Yeo, H. Chang, J. Song, K. Ahn, and V. N. Kim
Science
(2018), 361(6403):701–704
PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay
H. Yi, J. Park, M. Ha, J. Lim, H. Chang, and V. N. Kim
Molecular Cell
(2018), 70(6):1081–1088.e5
‣
Terminal Uridylyltransferases Execute Programmed Clearance of Maternal Transcriptome in Vertebrate Embryos
H. Chang1, J. Yeo1, J.-g. Kim, H. Kim, J. Lim, M. Lee, H. H. Kim, J. Ohk, H.-Y. Jeon, H. Lee, H. Jung, K.-W. Kim, and V. N. Kim
Molecular Cell
(2018), 70(1):72–82
mTAIL-seq Reveals Dynamic Poly(A) Tail Regulation in Oocyte-to-Embryo Development
J. Lim, M. Lee, A. Son, H. Chang, and V. N. Kim
Genes & Development
(2016), 30:1671–1682
Regulation of Poly(A) Tail and Translation During the Somatic Cell Cycle
J.-E. Park, H. Yi, Y. Kim, H. Chang, and V. N. Kim
Molecular Cell
(2016), 62(3):462-471
Next-Generation Libraries for Robust RNA Interference-Based Genome-Wide Screens
M. Kampmann, M. A. Horlbeck, Y. Chena, J. C. Tsai, M. C. Bassik, L. A. Gilbert, J. E. Villalta, S. C. Kwon, H. Chang, V. N. Kim, and J. S. Weissman
Proceedings of the National Academy of Sciences of the U. S. A.
(2015), 112(26):E3384-E3391
Temporal Landscape of MicroRNA-Mediated Host-Virus Crosstalk during Productive Human Cytomegalovirus Infection
S. Kim, D. Seo, D. Kim, Y. Hong, H. Chang, D. Baek, V. N. Kim, S. Lee, and K. Ahn
Cell Host & Microbe
(2015), 17(6):838–851
TUT7 Controls the Fate of Precursor MicroRNAs by Using Three Different Uridylation Mechanisms
B. Kim, M. Ha, L. Loeff, H. Chang, D. K. Simanshu, S. Li, M. Fareh, D. J. Patel, C. Joo, and V. N. Kim
EMBO Journal
(2015), 35(2):115-236
‣
miRseqViewer: Multi-Panel Visualization of Sequence, Structure and Expression for Analysis of MicroRNA Sequencing Data
I. Jang1, H. Chang1, Y. Jun, S. Park, J. O. Yang, B. Lee, W. Kim, V. N. Kim, and S. Lee
Bioinformatics
(2015), 31(4):596–598
‣
Uridylation by TUT4 and TUT7 Marks mRNA for Degradation
J. Lim1, M. Ha1, H. Chang1, S. C. Kwon, D. K. Simanshu, D. J. Patel, and V. N. Kim
Cell
(2014), 159(6):1365–1376
‣
TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications
H. Chang1, J. Lim1, M. Ha, and V. N. Kim
Molecular Cell
(2014), 53(6):1044–1052
‣
FitSearch: a Robust Way to Interpret a Yeast Fitness Profile in Terms of Drug’s Mode-of-Action
M. Lee1, S. Han1, H. Chang1, Y.-S. Kwak, D. M. Weller, and D. Kim
BMC Genomics
(2014), 14(Suppl 1):S6
Construction of the First Compendium of Chemical-Genetic Profiles in the Fission Yeast Schizosaccharomyces pombe and Comparative Compendium Approach
S. Han, M. Lee, H. Chang, M. Nam, H.-O. Park, Y.-S. Kwak, H.-J. Ha, D. Kim, S.-O. Hwang, K.-L. Hoe, and D.-U. Kim
Biochemical and Biophysical Research Communications
(2013), 436(4):613–618
miRGator v3.0: a MicroRNA Portal for Deep Sequencing, Expression Profiling and mRNA Targeting
S. Cho, I. Jang, Y. Jun, S. Yoon, M. Ko, Y. Kwon, I. Choi, H. Chang, D. Ryu, B. Lee, V. N. Kim, W. Kim, and S. Lee
Nucleic Acids Research
(2013), 41(D1):D252–D257
‣
LIN28A is a Suppressor of ER-associated Translation in Embryonic Stem Cells
J. Cho1, H. Chang1, S. C. Kwon, B. Kim, Y. Kim, J. Choe, M. Ha, Y. K. Kim, and V. N. Kim
Cell
(2012), 151(4):765–777
Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II Let-7 MicroRNAs
I. Heo, M. Ha, J. Lim, M.-J. Yoon, J.-E. Park, S. C. Kwon, H. Chang, and V. N. Kim
Cell
(2012), 151(3):521–532
Dicer recognizes the 5′ end of RNA for efficient and accurate processing
J.-E. Park, I. Heo, Y. Tian, D. K. Simanshu, H. Chang, D. Jee, D. J. Patel, and V. N. Kim
Nature
(2011), 475(7355):201-205

